Find Shared DNA
Shared DNA between two or more individuals can be identified with
find_shared_dna(). One PNG file and up to two CSV files are output when
save_output=True.
In the filenames below,
name1 is the name of the first Individual
name2 is the name of the second Individual
cM_threshold corresponds to the same named parameter of
find_shared_dna(); “.” is replaced by “p” with precision of 2, e.g., “0p75”
snp_threshold corresponds to the same named parameter of
find_shared_dna()
genetic_map corresponds to the same named parameter of
find_shared_dna().
Note
If more than two individuals are compared, all Individual
names will be included in the filenames and plot titles using the same conventions.
Note
Genetic maps do not have recombination rates for the Y chromosome since the Y
chromosome does not recombine. Therefore, shared DNA will not be shown on the Y
chromosome.
shared_dna_<name1>_<name2>_<cM_threshold>cM_<snp_threshold>snps_GRCh37_<genetic_map>.png
This plot illustrates shared DNA (i.e., no shared DNA, shared DNA on one chromosome, and shared
DNA on both chromosomes). The centromere for each chromosome is also detailed. Two examples of
this plot are shown below.
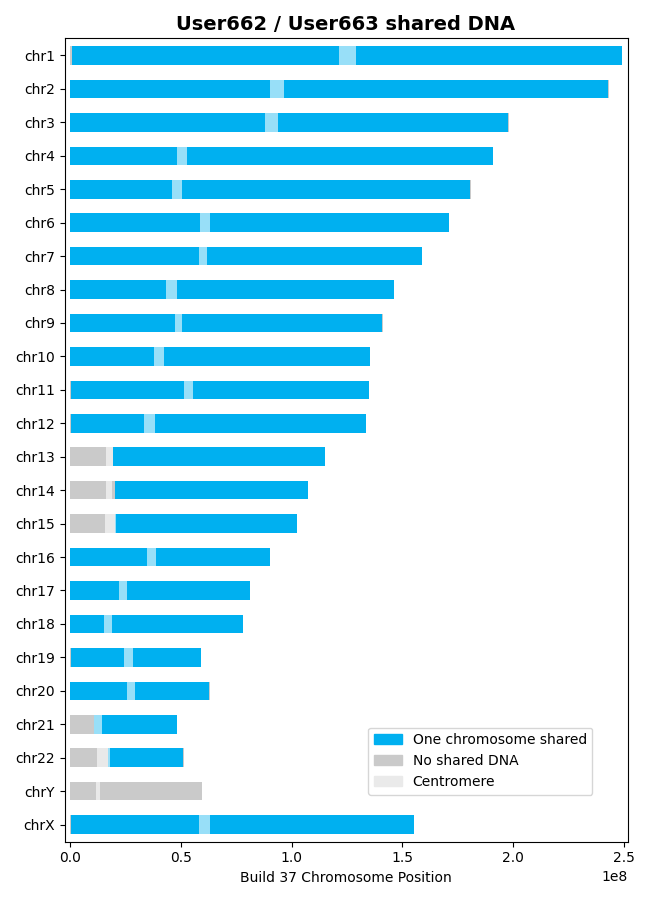
In the above plot, note that the two individuals only share DNA on one chromosome. In this plot,
the larger regions where “No shared DNA” is indicated are due to SNPs not being available in
those regions (i.e., SNPs were not tested in those regions).
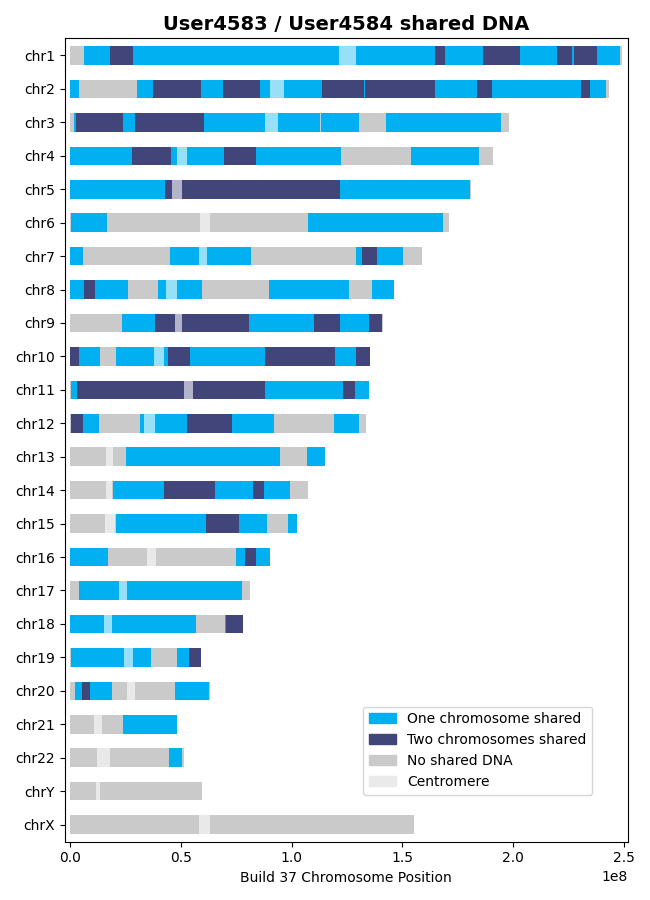
In the above plot, the areas where “No shared DNA” is indicated are the regions where SNPs were
not tested or where DNA is not shared. The areas where “One chromosome shared” is indicated are
regions where the individuals share DNA on one chromosome. The areas where “Two chromosomes
shared” is indicated are regions where the individuals share DNA on both chromosomes in the pair
(i.e., the individuals inherited the same DNA from their father and mother for those regions).
Note that the regions where DNA is shared on both chromosomes is a subset of the regions where
one chromosome is shared.
shared_dna_one_chrom_<name1>_<name2>_<cM_threshold>cM_<snp_threshold>snps_GRCh37_<genetic_map>.csv
If DNA is shared on one chromosome, a CSV file details the shared segments of DNA.
Column |
Description |
|---|
segment |
Shared DNA segment number |
chrom |
Chromosome with matching DNA segment |
start |
Start position of matching DNA segment |
end |
End position of matching DNA segment |
cMs |
CentiMorgans of matching DNA segment |
snps |
Number of SNPs in matching DNA segment |
shared_dna_two_chroms_<name1>_<name2>_<cM_threshold>cM_<snp_threshold>snps_GRCh37_<genetic_map>.csv
If DNA is shared on two chromosomes, a CSV file details the shared segments of DNA.
Column |
Description |
|---|
segment |
Shared DNA segment number |
chrom |
Pair of chromosomes with matching DNA segment |
start |
Start position of matching DNA segment on each chromosome |
end |
End position of matching DNA segment on each chromosome |
cMs |
CentiMorgans of matching DNA segment on each chromosome |
snps |
Number of SNPs in matching DNA segment on each chromosome |
Find Shared Genes
Shared genes (with the same genetic variations) between two or more individuals can be
identified with find_shared_dna(), with the parameter shared_genes=True.
In addition to the outputs produced by Find Shared DNA, up to two additional CSV files are
output that detail the shared genes when save_output=True.
In the filenames below, name1 is the name of the first
Individual and name2 is the name of the second
Individual. (If more individuals are compared, all
Individual names will be included in the filenames using the same
convention.)
shared_genes_one_chrom_<name1>_<name2>_<cM_threshold>cM_<snp_threshold>snps_GRCh37_<genetic_map>.csv
If DNA is shared on one chromosome, this file details the genes shared between the individuals
on at least one chromosome; these genes are located in the shared DNA segments specified in
shared_dna_one_chrom_<name1>_<name2>_<cM_threshold>cM_<snp_threshold>snps_GRCh37_<genetic_map>.csv.
Column* |
Description* |
|---|
name |
Name of gene |
geneSymbol |
Gene symbol |
chrom |
Reference sequence chromosome or scaffold |
strand |
+ or - for strand |
txStart |
Transcription start position (or end position for minus strand item) |
txEnd |
Transcription end position (or start position for minus strand item) |
refseq |
RefSeq ID |
proteinID |
UniProt display ID, UniProt accession, or RefSeq protein ID |
description |
Description |
* UCSC Genome Browser /
UCSC Table Browser

